(Updated 9/2/2009, but still unfinished; see other’s work on this that I’ve collected)
I never took a statistics class, so I only know the kind of statistics you learn on the street. But now that I’m in global health research, I’ve been doing a lot of on-the-job learning. This post is about something I’ve been reading about recently, how to decide if a simple statistical model is sufficient or if the data demands a more complicated one. To keep the matter concrete (and controversial) I’ll focus on a claim from a recent paper in Nature that my colleague, Haidong Wang, choose for our IHME journal club last week: Advances in development reverse fertility declines. The title of this short letter boldly claims a causal link between total fertility rate (an instantaneous measure of how many babies a population is making) and the human development index (a composite measure of how “developed” a country is, on a scale of 0 to 1). Exhibit A in their case is the following figure:
An astute observer of this chart might ask, “what’s up with the scales on those axes?” But this post is not about the visual display of quantitative information. It is about deciding if the data has a piecewise linear relationship that Myrskyla et al claim, and doing it in a Bayesian framework with Python and PyMC. But let’s start with a figure where the axes have a familiar linear scale!
I was able to produce this alternative plot without working much because the authors and the journal made their dataset easily available on the web as a csv file. Thanks, all! In fact, they have provided more data than 1975 and 2005. Here is everything from the years in between as well:
Since we discussed this in journal club, I can now repeat my fellow post-docs’ observations and sound smart. In fact, I already did with that bit about the axes being on a non-linear (and non-logarithmic) scale. The letter claims that there is a breakpoint around human development index .86, and the data is piecewise linear with decreasing slope below this value, and increasing above. The next observation that is not mine originally, is that the data also looks like a quadratic would fit it pretty well.
To be slightly more formal, here are two alternative models for the association between HDI and TFR:
and
How should I decide between these two models? Well, I’m open to suggestions, but I learned about one way when I read a citation classic by UW statistician Adrian Raftery and co-author about Bayes Factors. They describe the ratio . If K is more than 3.2, this constitutes “substantial” evidence that model 2 is superior.
Since I’ve titled this post in the form of a tutorial, I’m now going to go through calculating the Bayes factor with MCMC in Python, which turns out to be a slightly challenging computation (but easy to code; thanks, PyMC!).
To set up the models in the fully Bayesian way, we need some priors, which I’ve made up below in a convenient, reusable form:
class linear:
def __init__(self, X, Y, order=1, mu_beta=None, sigma_beta=1., mu_sigma=1.):
if mu_beta == None:
mu_beta = zeros(order+1)
self.beta = Normal('beta', mu_beta, sigma_beta**-2)
self.sigma = Gamma('standard error', 1., 1./mu_sigma)
@potential
def data_potential(beta=self.beta, sigma=self.sigma,
X=X, Y=Y):
mu = self.predict(beta, X)
return normal_like(Y, mu, 1 / sigma**2)
self.data_potential = data_potential
def predict(self, beta, X):
return polyval(beta, X)
def logp(self):
logp = []
for beta_val, sigma_val in zip(self.beta.trace(), self.sigma.trace()):
self.beta.value = beta_val
self.sigma.value = sigma_val
logp.append(Model(self).logp)
return array(logp)
and
class piecewise_linear:
def __init__(self, X, Y, breakpoint=.86,
mu_beta=[0., 0., 0.], sigma_beta=1., mu_sigma=1.):
self.breakpoint=breakpoint
self.beta = Normal('beta', mu_beta, sigma_beta**-2)
self.sigma = Gamma('standard error', 1., 1./mu_sigma)
@potential
def data_potential(beta=self.beta, sigma=self.sigma,
X=X, Y=Y, breakpoint=self.breakpoint):
mu = self.predict(beta, breakpoint, X)
return normal_like(Y, mu, 1 / sigma**2)
self.data_potential = data_potential
def predict(self, beta, breakpoint, X):
very_high_dev_indicator = X >= breakpoint
mu = (beta[0] + beta[1]*(X-breakpoint)) * (1 - very_high_dev_indicator)
mu += (beta[0] + beta[2]*(X-breakpoint)) * very_high_dev_indicator
return mu
def logp(self, beta_val=None, sigma_val=None, breakpoint_val=None):
for beta_val, sigma_val in zip(self.beta.trace(), self.sigma.trace()):
self.beta.value = beta_val
self.sigma.value = sigma_val
logp.append(Model(self).logp)
return array(logp)
Then to try to calculate the Bayes factor, I can draw samples from the posterior distribution of each model, and look at the harmonic mean of their posterior liklihoods.
def bayes_factor(m1, m2, iter=1e6, burn=25000, thin=10, verbose=0):
MCMC(m1).sample(iter, burn, thin, verbose=verbose)
logp1 = m1.logp()
MCMC(m2).sample(iter, burn, thin, verbose=verbose)
logp2 = m2.logp()
mu_logp = mean(logp2)
K = exp(pymc.flib.logsum(-logp1) - log(len(logp1))
- (pymc.flib.logsum(-logp2) - log(len(logp2))))
return K
Unfortunately, it seems to take a prohibitively large number of samples to get the same average twice.
To make it all run, I’ve made a little module to load the data, but I won’t bore you with the details; it’s online here if you want to play around with it yourself.
>>> import data >>> m1=models.linear(X=data.hdi, Y=data.tfr, order=2) >>> m2=models.piecewise_linear(X=data.hdi, Y=data.tfr, mu_beta=[1,-10,1]) >>> model_selection.bayes_factor(m1, m2, iter=1e7)
According to the Bayes factor, the piecewise linear model is (to be filled in soon) better than the quadratic model. Or, more quantitatively, the observed data is (tba) times more likely under model 2 than model 1. Cool!
As a side-effect, this yields an alternative way to ask if the association on the “high development” piece of the piecewise model is really positive:
>>> m2 = models.piecewise_linear(X=data.hdi, Y=data.tfr, breakpoint=.86, mu_beta=[1,-8,1], sigma_beta=1., mu_sigma=.1)
>>> MCMC(m2).sample(iter=1000*1000+20000, thin=1000, burn=20000, verbose=1)
>>> m2.beta.stats()['95% HPD interval']
array([[ 1.91520875, 2.01669476],
[-9.84036014, -9.48869047],
[-3.85410676, -1.23869569]])
>>> m2.beta.stats()['mean']
array([ 1.96932651, -9.66349812, -2.5683748 ])
The research questions for the computer scientist in me are: did I draw enough samples to get a correct answer? and did I really need to draw that many?
The tentative answers are no and no! See the comments for leads on more efficient schemes.




Oh, wikipedia was very helpful to me on this:
http://en.wikipedia.org/wiki/Bayes_factor
What? A Nature paper overselling its results?
NEVAR!!!
Nice.
Since you can call R functions from Python (rpy , RSPython), there are multiple ways to do MCMC in Python
Aren’t the axes logarithmic? The Y axis looks conventionally so, and the X axis might be if you take 1-X and look from the right. Evidently full development is something that can only be approached, never achieved.
If you determine that you drew more samples than you needed, can you get a refund?
Hi Abie,
Thanks for another interesting post! One minor point is that you compute Bayes’ factors by averaging the likelihood with respect to the prior:
whereas what you’ve got is
So to compute a Bayes’ factor, you don’t need to run an MCMC, you just need to sample from the prior.
This is surprising at first, but makes sense if you think about it: models that ‘inherently’ predict the data are preferred. The “Occam’s razor effect”, ie Bayesian statistics’ empirical preference for simple models, is a consequence. Simple models’ predictions are generally less variable than complicated models’, so if a dataset is supported by a simple model it’s probably relatively likely wrt that model also.
In PyMC, you can draw samples from the prior using the method Model.draw_from_prior(). There’s a package called MultiModelInference in the sandbox that will compute Bayes’ factors, too.
MultiModelInference is in the sandbox because it’s really hard to guarantee reasonable performance in most practical cases. Estimates of Bayes’ factors by Monte Carlo integration just get really bad as model complexity increases. The mode of failure is that one sampled parameter set ends up with a vastly higher likelihood than any other. This is the main reason for the popularity of alternatives to Bayes’ factors, such as DIC.
Maria: Right?
efrique: Thanks, and good point. I’ve been meaning to figure out how to use rpy; then I could overlay the loess smoothed version of the data above on my linear-scale version of the paper plot. Anyone want to show me how?
Greg: I haven’t seen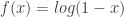 as a “log scale” before, but it makes sense for data that can’t go above 1, as does
as a “log scale” before, but it makes sense for data that can’t go above 1, as does 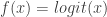 . Maybe that’s what’s going on here. In light of Anand’s comments, you should get a refund for the sampled quantity being defective!
. Maybe that’s what’s going on here. In light of Anand’s comments, you should get a refund for the sampled quantity being defective!
Anand: Thanks for that clarification! I’ll update this tutorial as soon as I have another break. What you say makes sense, but leaves me wondering if there is a sneaky way to use MCMC to circumvent the problem of MC in complex models. Let me try, see if you buy it:

 ?
?
so is what I calculated
Anand: I think I figured out the MCMC way to do it:
![E[f(\theta)] = \int f(\theta) p(\theta|data, M) = \int f(\theta) p(\theta, data | M) / p(data | M)](https://s0.wp.com/latex.php?latex=E%5Bf%28%5Ctheta%29%5D+%3D+%5Cint+f%28%5Ctheta%29+p%28%5Ctheta%7Cdata%2C+M%29+%3D+%5Cint+f%28%5Ctheta%29+p%28%5Ctheta%2C+data+%7C+M%29+%2F+p%28data+%7C+M%29&bg=ffffff&fg=333333&s=0&c=20201002) so
so ![p(data | M) = \int f(\theta) p(\theta, data | M) / E[f(\theta)]](https://s0.wp.com/latex.php?latex=p%28data+%7C+M%29+%3D+%5Cint+f%28%5Ctheta%29+p%28%5Ctheta%2C+data+%7C+M%29+%2F+E%5Bf%28%5Ctheta%29%5D&bg=ffffff&fg=333333&s=0&c=20201002) . If we pick
. If we pick  , the numerator goes away. This might be due to Newton and Raftery originally.
, the numerator goes away. This might be due to Newton and Raftery originally.
So to me the critical issue here is sensitivity to the choice of the hyperparameters in the beta prior you’re using. Obviously this can potentially affect the results.
I don’t really understand how the posterior for \beta_3 could have negative mean. The prior was zero mean, and that data clearly slopes upward.
Thinking about it further, I think you need to be more careful in your interaction between your model specification and your choice of prior.
Thought experiment for the piecewise linear model. Keep your same data, and your same model, but add 100 to all your X values. What’s going to happen?
I think that if you’re going to fit piecewise linear models where you already know the breakpoints (which is a little iffy to begin with, isn’t it?) and you’re putting the same prior on the coefficient of each piece, you need to do some normalization.
Good idea, I hadn’t thought of or come across that before. It says here: http://reference.kfupm.edu.sa/content/h/y/hypothesis_testing_and_model_selection_v_1311334.pdf that that method suffers from a similar problem, because if is really small for some posterior samples, those samples will dominate the harmonic mean.
is really small for some posterior samples, those samples will dominate the harmonic mean.
However, there are lots of ideas for improvements in that paper, and it’s pretty old. Maybe there’s hope for MultiModelInference after all!
rif: Say, is this the rif from Harvey Mudd?
rif: agreed re fixing the breakpoint. I tried to write the code in a way that makes it easy to add a stochastic breakpoint also, with e.g.
piecewise_linear(X=data.hdi, Y=data.tfr, breakpoint=Normal('bp', .86, 100)), but I didn’t test if it actually works.Re the slope upwards, I want to be clear that I am looking at all country-years, i.e. the slope of the blue, red, and grey dots right on .86 on https://healthyalgorithms.files.wordpress.com/2009/08/hdi_v_tfr31.png
Re the priors, your thought experiment is a good one. But I know I _can_ find priors for which the posterior \beta_3 is positive, like N(10,1). Is there an “objective” way to renormalize?
Anand: I have *heard* of rif from Harvey Mudd before! But I am not him! I am rif from MIT. I am not such a fount of Random Information. I would like to meet rif from HM though. He doesn’t seem to be too easily findable online.
Abie: I guess for all the points, it’s a lot harder to tell whether there’s a slope there or not. It’d be at least amusing to look at just that part of the graph, blown up.
My point is that with the prior you’re using, if we take some data far out on the x axis and close to the y axis with an obvious upward slope, it is going to be “more likely” to fit (intercept = average, slope = 0) then (intercept = big negative number, slope = moderate positive number) because your beta prior makes that big negative number very unlikely. Your conclusions are only as good as your prior is plausible, and in this case, it’s easy to see that your prior is heavily constraining the solution away from nonzero slopes.
You’re the Bayesian statistician and I’m just a lowly software engineer, so I probably don’t have to tell you that choosing the prior is the hard part. There is no objective way to normalize. Choosing the prior is the entire art. In this case, since you’re already considering fixed breakpoints, I might consider a model where you only fit slopes, and normalized the data points in each linear piece to have mean 0 in both x and y.
Of course, when I think of a piecewise linear function, I usually think of a piecewise linear continuous function, so you also could think about enforcing that somehow.
In any case, with the computation as it stands, I don’t really buy your conclusion about the slope.
rif: I was also thinking of a piecewise linear continuous function… but that’s not what I coded. Oops! Yet another lesson to me to verify predictions graphically, which would have shown me just what my model was saying.
I’m planning to update this whole business once I have a reliable estimate of the Bayes factor. But if you look on github now, you can see my new attempt. The piecewise linear model is continuous now, and I exposed more of the priors for easy experimentation.
I’ll have to think more about rescaling the data… what I’m leaning towards now is picking the priors to try to convince you: since you believe that the slope is positive, let’s make the prior positive. How positive does the prior need to be before the 95% uncertainty interval is all positive?
Well, it’s not just the prior on the slope, but also the prior on the intercept. I’m fine with a zero mean prior on the slope, if the prior on the intercept is wide enough that that the intercept value implied by a reasonable positive slope is plausible. So for instance, if the slope were 1 starting at x = 10, your y intercept is -10, so you’d better have a prior where -10 isn’t way out in the tails.
Have you tried Skilling’s Nested sampling?
rif: I think that some dialogue like this is a good way to select priors. I will make some pictures designed to convince you (just for fun… my main goal in this post is to figure out how to compute and use the Bayes factor).
Joe: I have not. Is it the method described in Nested sampling for general Bayesian computation? Now that I am approximating K correctly, I have to draw way more samples from the posterior than I would like. Maybe nested sampling is the remedy.
Yes thats the one. Its not entirely clear to me when this beats dumb sampling, but it is designed for this purpose.
Abie: see http://gist.github.com/179657 for a better way to sum probabilities on the log scale.
Nested sampling looks promising. It might be a bit complicated to implement, though, due to the need to run several sequential MCMC’s within the likelihood shells.
Does anyone understand why Skilling’s eq. 14 works? In particular, why doesn’t the distribution of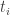 involve the shapes of the likelihood shells? In the
involve the shapes of the likelihood shells? In the 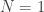 case, say?
case, say?
Anand: Thanks for the
pymc.flib.logsum. Maybe that will resolve some of the imprecision in my mean inverse liklihood, which are currently not very precise, even with many, many samples.I think that Skilling’s eq. 14 is so simple because all of the complexity is pushed off into the subroutine that samples from the prior conditioned on the likelihood being above some threshold.
One way to implement this in PyMC would be a potential that has a parameter which has no step method, and is changed manually at each iteration of the nested sampling.
Hi Abie,
interesting post and comments. Just a few questions:
– when you fit the piecewise linear to all country-years available, what’s the size of beta_2 and is it significantly >0?
– what time paths emerge if you pick some high-income OECD country (e.g., UK), some middle-income country (e.g., Argentinia, Brazil), and some low-income country (e.g., Malawi)?
The additive, weighted calculation the HDI allows (partial) substitution of poor results in one area with good results in another (the inherent correlation structure in the components makes that a bit harder in practice than in theory, but let’s not get started on the good and bad of the HDI methodology). So when the turning point of 0.86 is chosen, it’s possible to surpass that in theory with gross enrollment of 0%, or literacy rate of 50%, or GDP of $10000 (not all 3 at the same time!). So one thought I have is that it’s not so much a reversal of fertility at higher stages of development but more countries joining the club and those newcomers have, albeit lower than in the past, fertility rates that are higher than those of the old club members. The democratization of wealth…
Tanja
Pingback: Holiday reading « Healthy Algorithms
I still have plans to work on this some more, but I also still do not have time.
Tanja: I’ll make some graphs like this when I make the zoomed graphs for Rif. I predict that the beta_2 95% HPD region will include 0, but I need to spend some more time to be sure.
I did have time to learn a little bit more about the culture wars surrounding these numbers and their interpretations, in The Means of Reproduction by Michelle Goldberg.
Pingback: Inequality vs Stuff « Healthy Algorithms
Pingback: MCMC in Python: Statistical model stuck on a stochastic system dynamics model in PyMC | Healthy Algorithms
Pingback: 2010 in review | Healthy Algorithms
Hi – the URL to the Krass_1995.pdf file is actually
ftp://ftp.stat.berkeley.edu/pub/users/binyu/212A/papers/Kass_1995.pdf
The URL posted above
ftp://stat-ftp.berkeley.edu/pub/users/binyu/212A/papers/Kass_1995.pdf
doesn’t work (at least from outside of UCB.)
Pingback: That Bayes Factor | Healthy Algorithms
@Cinead: Thanks, link updated.
Pingback: PyMC at SciPy 2011 | Healthy Algorithms